Run a SVM GMM system on the RSR2015 database¶
This script run an experiment on the male evaluation part of the RSR2015 database. The protocol used here is based on the one described in [Larcher14]. In this version, we only consider the non-target trials where impostors pronounce the correct text (Imp Correct).
The number of Target trials performed is then - TAR correct: 10,244 - IMP correct: 573,664
[Larcher14] Anthony Larcher, Kong Aik Lee, Bin Ma and Haizhou Li, “Text-dependent speaker verification: Classifiers, databases and RSR2015,” in Speech Communication 60 (2014) 56–77
Input/Output¶
Enter:¶
the number of distribution for the Gaussian Mixture Models
the root directory where the RSR2015 database is stored
Generates the following outputs:¶
a Mixture in compressed pickle format (ubm)
a StatServer of zero and first-order statistics (enroll_stat)
a StatServer of zero and first-order statistics (back_stat)
a StatServer of zero and first-order statistics (nap_stat)
a StatServer of zero and first-order statistics (test_stat)
a StatServer containing the super vectors of MAP adapted GMM models for each speaker (enroll_sv)
a StatServer containing the super vectors of MAP adapted GMM models for each speaker (back_sv)
a StatServer containing the super vectors of MAP adapted GMM models for each speaker (nap_sv)
a StatServer containing the super vectors of MAP adapted GMM models for each speaker (test_sv)
a score file
a DET plot
import numpy as np
import sidekit
import multiprocessing
import os
import sys
import matplotlib.pyplot as mpl
import logging
logging.basicConfig(filename='log/rsr2015_svm-gmm.log',level=logging.DEBUG)
Set your own parameters¶
distrib_nb = 512 # number of Gaussian distributions for each GMM
NAP = True # activate the Nuisance Attribute Projection
nap_rank = 40
rsr2015Path = '/lium/corpus/vrac/RSR2015_V1/'
# Set the number of parallel process to run.
nbThread = 10
Load IdMap, Ndx, Key from HDF5 files and ubm_list¶
that define the task. Note that these files are generated when running
rsr2015_init.py:
logging.info('Load task definition')
enroll_idmap = sidekit.IdMap('task/3sesspwd_eval_m_trn.h5')
nap_idmap = sidekit.IdMap('task/3sess-pwd_eval_m_nap.h5')
back_idmap = sidekit.IdMap('task/3sess-pwd_eval_m_back.h5')
test_ndx = sidekit.Ndx('task/3sess-pwd_eval_m_ndx.h5')
test_idmap = sidekit.IdMap('task/3sess-pwd_eval_m_test.h5')
key = sidekit.Key('task/3sess-pwd_eval_m_key.h5')
with open('task/ubm_list.txt') as inputFile:
ubmList = inputFile.read().split('\n')
Process the audio to save MFCC on disk¶
logging.info("Initialize FeaturesExtractor")
extractor = sidekit.FeaturesExtractor(audio_filename_structure=audioDir+"/{}.wav",
feature_filename_structure="./features/{}.h5",
sampling_frequency=16000,
lower_frequency=133.3333,
higher_frequency=6955.4976,
filter_bank="log",
filter_bank_size=40,
window_size=0.025,
shift=0.01,
ceps_number=19,
vad="snr",
snr=40,
pre_emphasis=0.97,
save_param=["vad", "energy", "cep"],
keep_all_features=False)
# Get the complete list of features to extract
show_list = np.unique(np.hstack([ubmList, enroll_idmap.rightids, np.unique(test_ndx.segset)]))
channel_list = np.zeros_like(show_list, dtype = int)
logging.info("Extract features and save to disk")
extractor.save_list(show_list=show_list,
channel_list=channel_list,
num_thread=nbThread)
Create a FeaturesServer¶
From this point, all objects that need to process acoustic features will do it through a FeaturesServer. This object is initialized here. We define the type of parameters to load (log-energy + cepstral coefficients) and the post-process to apply on the fly (RASTA filtering, CMVN, addition iof the first and second derivatives, feature selection).
# Create a FeaturesServer to load features and feed the other methods
features_server = sidekit.FeaturesServer(features_extractor=None,
feature_filename_structure="./features/{}.h5",
sources=None,
dataset_list=["energy", "cep", "vad"],
mask=None,
feat_norm="cmvn",
global_cmvn=None,
dct_pca=False,
dct_pca_config=None,
sdc=False,
sdc_config=None,
delta=True,
double_delta=True,
delta_filter=None,
context=None,
traps_dct_nb=None,
rasta=True,
keep_all_features=False)
Train the Universal background Model (UBM)¶
An empty Mixture is initialized and an EM algorithm is run to estimate the UBM before saving it to disk. Covariance matrices are diagonal in this example.
logging.info('Train the UBM by EM')
# load all features in a list of arrays
ubm = sidekit.Mixture()
llk = ubm.EM_split(features_server,
ubmList,
distrib_nb,
num_thread=nbThread)
ubm.write('gmm/ubm.h5')
Compute the sufficient statistics on the UBM¶
Make use of the new UBM to compute the sufficient statistics of all enrolement sessions that should be used to train the speaker GMM models, models for the SVM training blacklist, segments to train the NAP matrix and test segments. An empty StatServer is initialized. Statistics are then computed in the StatServer which is then stored to disk:
logging.info()
enroll_stat = sidekit.StatServer(enroll_idmap,
distrib_nb=512,
feature_size=60)
enroll_stat.accumulate_stat(ubm=ubm,
feature_server=features_server,
seg_indices=range(enroll_stat.segset.shape[0]),
num_thread=nbThread)
enroll_stat.write('data/stat_rsr2015_male_enroll.h5')
back_stat = sidekit.StatServer(back_idmap,
distrib_nb=512,
feature_size=60)
back_stat.accumulate_stat(ubm=ubm,
feature_server=features_server,
seg_indices=range(back_stat.segset.shape[0]),
num_thread=nbThread)
back_stat.write('data/stat_rsr2015_male_back.h5')
nap_stat = sidekit.StatServer(nap_idmap,
distrib_nb=512,
feature_size=60)
nap_stat.accumulate_stat(ubm=ubm,
feature_server=features_server,
seg_indices=range(nap_stat.segset.shape[0]),
num_thread=nbThread)
nap_stat.write('data/stat_rsr2015_male_nap.h5')
test_stat = sidekit.StatServer(test_idmap,
distrib_nb=512,
feature_size=60)
test_stat.accumulate_stat(ubm=ubm,
feature_server=features_server,
seg_indices=range(test_stat.segset.shape[0]),
num_thread=nbThread)
test_stat.write('data/stat_rsr2015_male_test.h5')
Train a GMM for each session¶
Only adapt the mean super-vector and store all of them in the enrol_sv StatServer that is then stored in compressed picked format:
logging.info('MAP adaptation of the speaker models')
regulation_factor = 3 # MAP regulation factor
enroll_sv = enroll_stat.adapt_mean_map(ubm, regulation_factor, norm=True)
enroll_sv.write('data/sv_norm_rsr2015_male_enroll.h5')
back_sv = back_stat.adapt_mean_map(ubm, regulation_factor, norm=True)
back_sv.write('data/sv_rsr2015_male_back.h5')
nap_sv = nap_stat.adapt_mean_map(ubm, regulation_factor, norm=True)
nap_sv.write('data/sv_rsr2015_male_nap.h5')
test_sv = test_stat.adapt_mean_map(ubm, regulation_factor, norm=True)
test_sv.write('data/sv_rsr2015_male_test.h5')
Apply Nuisance Attribute Projection if required¶
If NAP == True, estimate and apply the Nuisance Attribute Projection
on all supervectors:
if NAP:
logging.info('Estimate and apply NAP')
napMat = back_sv.get_nap_matrix_stat1(nap_rank);
back_sv.stat1 = back_sv.stat1 - np.dot(np.dot(back_sv.stat1, napMat), napMat.transpose())
enroll_sv.stat1 = enroll_sv.stat1 - np.dot(np.dot(enroll_sv.stat1, napMat), napMat.transpose())
test_sv.stat1 = test_sv.stat1 - np.dot(np.dot(test_sv.stat1, napMat), napMat.transpose())
Train the Support Vector Machine models¶
Train a Support Vector Machine for each speaker by considering the three sessions of this speaker:
logging.info('Train the SVMs')
sidekit.svm_training('svm/', back_sv, enroll_sv, num_thread=nbThread)
Compute all trials and save scores in HDF5 format¶
Compute the scores for all trials:
logging.info('Compute trial scores')
scores_gmm_svm = sidekit.svm_scoring('svm/{}.svm', test_sv, test_ndx, num_thread=nbThread)
if NAP:
scores_gmm_svm.write('scores/scores_svm-gmm_NAP_rsr2015_male.h5')
else:
scores_gmm_svm.write('scores/scores_svm-gmm_rsr2015_male.h5')
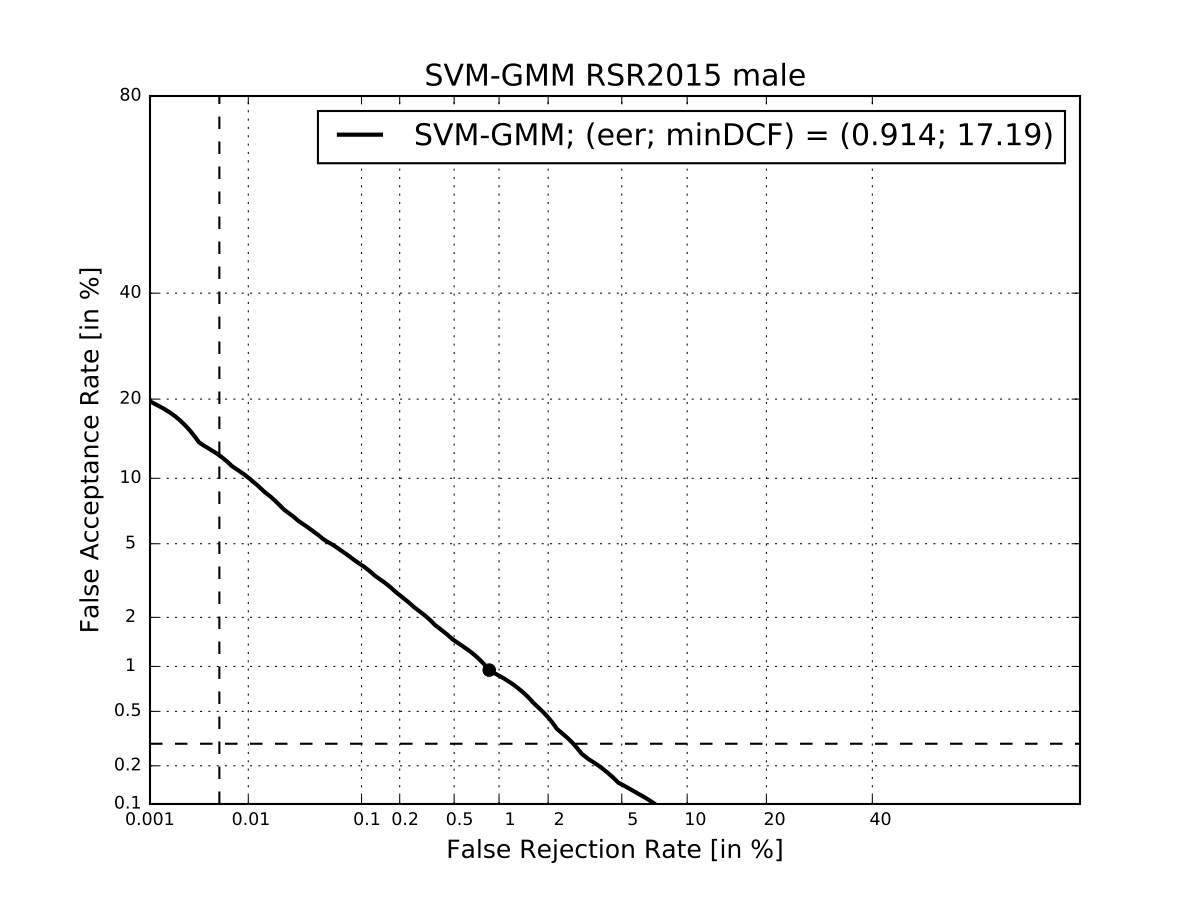
Plot DET curve and compute minDCF and EER¶
logging.info('Plot the DET curve')
prior = sidekit.logit_effective_prior(0.01, 10, 1)
# Initialize the DET plot to 2008 settings
dp = sidekit.DetPlot(window_style='sre10', plot_title='SVM-GMM RSR2015 male')
dp.set_system_from_scores(scores_gmm_svm, key, sys_name='SVM-GMM')
dp.create_figure()
dp.plot_rocch_det(0)
dp.plot_DR30_both(idx=0)
dp.plot_mindcf_point(prior, idx=0)
minDCF, Pmiss, Pfa, prbep, eer = sidekit.bosaris.detplot.fast_minDCF(dp.__tar__[0], dp.__non__[0], prior, normalize=True)
logging.info("minDCF = {}, eer = {}".format(minDCF, eer))
After running this script you should obtain the following curve¶