Diarization for ASR¶
This script performs a BIC diarization (ussally for ASR decoding)
The proposed diarization system was inspired by the system~:raw-latex:cite{Barras06} which won the RT‘04 fall evaluation and the ESTER,1 evaluation. It was developed during the ESTER,2 evaluation campaign for the transcription with the goal of minimizing word error rate.
Automatic transcription requires accurate segment boundaries. Segment boundaries have to be set within non-informative zones such as filler words.
Speaker diarization needs to produce homogeneous speech segments; however, purity and coverage of the speaker clusters are the main objectives here. Errors such as having two distinct clusters (i.e., detected speakers) corresponding to the same real speaker, or conversely, merging segments of two real speakers into only one cluster, get heavier penalty in the NIST time-based diarization metric than misplaced boundaries~:raw-latex:cite{NIST2004a}.
The system is composed of acoustic BIC segmentation followed with BIC hierarchical clustering. Viterbi decoding is performed to adjust the segment boundaries.
Music and jingle regions are not removed but a speech activity diarization could be load before to segment and cluster the show.
Optionally, long segments are cut to be shorter than 20 seconds.
%matplotlib inline
__license__ = "LGPL"
__author__ = "Sylvain Meignier"
__copyright__ = "Copyright 2015-2016 Sylvain Meignier"
__license__ = "LGPL"
__maintainer__ = "Sylvain Meignier"
__email__ = "sidekit@univ-lemans.fr"
__status__ = "Production"
__docformat__ = 'reStructuredText'
import argparse
import logging
import matplotlib
import copy
from matplotlib import pyplot as plot
from s4d.utils import *
from s4d.diar import Diar
from s4d import viterbi, segmentation
from sidekit import features_server
from s4d.clustering import hac_bic
from sidekit.sidekit_io import init_logging
from s4d.gui.dendrogram import plot_dendrogram
BIC diarization¶
Arguments, variables and logger¶
Set the log level
loglevel = logging.INFO
Set the input audio or mfcc file and the speech activity detection file (here not set).
data_dir = 'data'
show='20041219_1300_1314_RTM_ELDA'
input_show = os.path.join(data_dir, 'audio',show+'.sph')
#input_sad = None
input_sad = os.path.join(data_dir, 'sns',show+'.sns.seg')
Size of left or right windows (step 2)
win_size=250
Threshold for: * linear segmentation (step 3) * BIC HAC (step 4) * Vitterbi (step 5)
thr_l = 2
thr_h = 3
thr_vit = -250
If save_all is True then all produced diarization are saved
save_all = True
Set the logger options: logge information in console and file, set the level.
init_logging( level=loglevel)
Check if we work with an audio file or a mffc in spro4 format
input_fn = path_show_ext(input_show)
ffile = 'spro4'
if input_fn[2] in ['.sph', '.wav']:
ffile = 'audio'
logging.info('type of input: '+ffile)
2015-12-09 13:41:52,909 - root - INFO - type of input: audio
Prepare various variables
wdir = os.path.join('out', show)
if not os.path.exists(wdir):
os.makedirs(wdir)
Step 0 : MFCC¶
Get a Feature server instance
logging.info('Make MFCC')
fs = features_server.FeaturesServer(input_dir=input_fn[0],
input_file_extension=input_fn[2],
from_file=ffile,
config='diar_16k')
2015-12-09 13:41:52,916 - root - INFO - Make MFCC
Load MFCC or compute MFCC from audio
logging.info('Load show: %s', show)
tcep, vad = fs.load(show)
cep = tcep[0]
2015-12-09 13:41:52,920 - root - INFO - Load show: 20041219_1300_1314_RTM_ELDA
2015-12-09 13:41:52,921 - root - INFO - read audio
2015-12-09 13:41:53,069 - root - INFO - size of signal: 53.790894 len 14100960 type size 4
2015-12-09 13:41:53,666 - root - INFO - process part : 0.000000 881.310000 881.310000
2015-12-09 13:41:54,989 - root - INFO - no vad
2015-12-09 13:41:55,314 - root - INFO - keep log_e
2015-12-09 13:41:55,322 - root - INFO - !! size of signal cep: 8.740822 len 88129 type size 104
2015-12-09 13:41:55,327 - root - INFO - Smooth the labels and fuse the channels if more than one
2015-12-09 13:41:55,732 - root - INFO - no norm
Save the MFCC in spro4 format
mfcc_filename = os.path.join(wdir, show + '.pmfcc')
fs.save(show, mfcc_filename, 'spro4')
2015-12-09 13:41:55,737 - root - INFO - save spro4 format: out/20041219_1300_1314_RTM_ELDA/20041219_1300_1314_RTM_ELDA.pmfcc
Step 1¶
The initial diarization is loaded from a speech activity detection diarization (SAD) or a segment from the fist MFCC feature to the last MFCC feature is created.
logging.info('Check MFCC')
if input_sad is not None:
init_diar = Diar.read_seg(input_sad)
init_diar.pack(50)
else:
init_diar = seg.init_seg(cep, show)
if save_all:
init_filename = os.path.join(wdir, show + '.i.seg')
Diar.write_seg(init_filename, init_diar)
2015-12-09 13:41:55,983 - root - INFO - Check MFCC
Step 2: gaussian divergence segmentation¶
First segmentation: Segment each segment of init_diar using the
Gaussian Divergence method
logging.info('Make segmentation:')
#process every segment
seg_diar = Diar()
for seg in init_diar:
l = seg.length()
logging.debug('start: ', seg['start'],'end: ', seg['stop'], 'len: ', l)
if l > 2*win_size:
cep_seg = seg.seg_features(cep)
tmp = segmentation.div_gauss(cep_seg, show=show, win=win_size, shift=seg['start'])
seg_diar.append_diar(tmp)
else:
seg_diar.append_seg(seg)
i=0
for seg in seg_diar:
seg['label'] = 'S'+str(i)
i += 1
if save_all:
seg_filename = os.path.join(wdir, show + '.s.seg')
Diar.write_seg(seg_filename, seg_diar)
2015-12-09 13:41:55,998 - root - INFO - Make segmentation:
Step 3: linear BIC segmentation¶
This segmentation over the signal fuses consecutive segments of the same
speaker from the start to the end of the record. The measure employs the
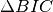 based on Bayesian Information Criterion , using full
covariance Gaussians (see class
based on Bayesian Information Criterion , using full
covariance Gaussians (see class gauss.GaussFull).
bicl_diar = segmentation.bic_linear(cep, seg_diar, thr_l, sr=False)
if save_all:
bicl_filename = os.path.join(wdir, show + '.l.seg')
Diar.write_seg(bicl_filename, bicl_diar)
2015-12-09 13:41:56,263 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,263 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,265 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,274 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,280 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,281 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,282 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,284 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,286 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,287 - root - WARNING - there is a hole between segment
2015-12-09 13:41:56,290 - root - WARNING - there is a hole between segment
147
Step 4: BIC HAC¶
Perform a BIC HAC
logging.info('Make clustering alpha: %f', thr_h)
bic = hac_bic.HAC_BIC(fs, bicl_diar, alpha=thr_h, sr=False)
bich_diar = bic.perform(to_the_end=True)
if save_all:
bichac_filename = os.path.join(wdir, show + '.h.seg')
Diar.write_seg(bichac_filename, bich_diar)
link, data = plot_dendrogram(bic.merge, 0, size=(25,6), log=True)
2015-12-09 13:41:56,298 - root - INFO - Make clustering alpha: 3.000000
/Users/meignier/pyenv3/lib/python3.5/site-packages/matplotlib/collections.py:590: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
if self._edgecolors == str('face'):
Step 4: re-segmentation¶
Viterbi decoding * HMM is trained: one GMM per speaker, GMM has 8 component with diagonal covariance matrix. Only a penalty between state is fixed. * Emission is computer: likelyhood for each feature * a Viterbi decoding is performed
logging.info('Make viterbi penalties: %f', thr_vit)
hmm = viterbi.Viterbi(cep, bich_diar, exit_penalties=[thr_vit])
hmm.train()
hmm.emission()
vit_diar = hmm.decode(init_diar)
if save_all:
vit_filename = os.path.join(wdir, show + '.d.seg')
Diar.write_seg(vit_filename, vit_diar)
2015-12-09 13:41:56,734 - root - INFO - Make viterbi penalties: -250.000000
Step 5¶
Move the segment boundaries into low energy aeras.
# adj_table = seg.adjust(cep, vit_table)
# if save_all:
# Table.write_seg(adj_filename, adj_table)
# output_seg(args.output, adj_table)
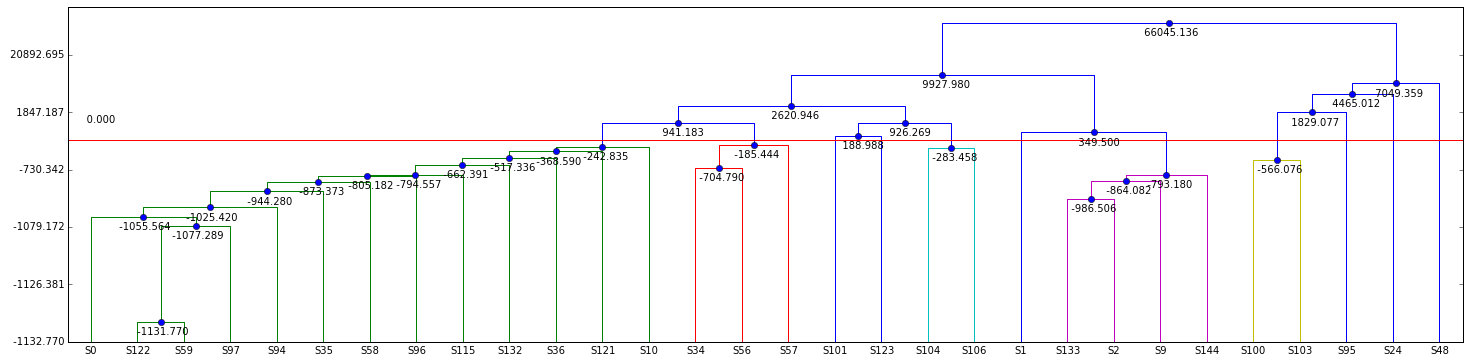
Compute the diarization error rate¶
from s4d.scoring import DER
from s4d.gui.viewer import PlotDiar
from s4d.gui.viewer_utils import diar_diff
ref = Diar.read_seg(os.path.join(data_dir, 'ref', show + '.seg'))
uem = Diar.read_uem(os.path.join(data_dir, 'ref', show + '.uem'))
der = DER(vit_diar, ref, uem, collar=25, no_overlap=False)
der.confusion()
der.assignment()
res = der.error()
print(res.rate_header())
print(res.time())
print(res.rate())
diff_diar = diar_diff(vit_diar, ref)
p = PlotDiar(diff_diar, size=(25, 6))
p.draw()
|| sns fa miss | speaker fa miss conf ||
|| 1762.62s 12.49s 3.89s | 827.56s 12.49s 3.89s 254.23s ||
|| 0.93% 0.71% 0.22% | 32.70% 1.51% 0.47% 30.72% ||
uem from ref
append collar
/Users/meignier/pyenv3/lib/python3.5/site-packages/matplotlib/figure.py:1653: UserWarning: This figure includes Axes that are not compatible with tight_layout, so its results might be incorrect.
warnings.warn("This figure includes Axes that are not "
Speaker diarization¶
MFCC for Speaker clustering¶
- get 12 MFCC + Delta and normalize them
from sidekit.frontend.features import compute_delta
from s4d.utils import cep_sliding_norm
#print('baseline cep:', cep.shape)
cep12 = cep[:,1:]
#print('12 MFCC:', cep12.shape)
delta = compute_delta(cep12)
#print('12 delta:', delta.shape)
cep24 = np.column_stack((cep12, delta))
#print('12MFCC + 12 delta:', cep24.shape)
cep_sliding_norm(cep24, win=301, center=True, reduce=True)
#hack put the new cep24 in the feature server
fs.cep[0] = cep24
#print(fs.cep.shape)
CLR clustering¶
- load UBM
from sidekit.mixture import Mixture
ubm = Mixture()
ubm_fn = os.path.join(data_dir, 'model', 'ubm128.gmm')
print(ubm_fn)
ubm.read_pickle(ubm_fn)
data/model/ubm128.gmm
- perform CLR HAC clustreing
- initialize HAC
- compute models
- compute distance
- do clustreing
from s4d.clustering.hac_clr import HAC_CLR
thr_clr = -1.4
clr_hac = HAC_CLR(fs, vit_diar, ubm)
clr_hac.initial_models()
clr_hac.initial_distances()
Warning, some arguments are not named, computation might not be parallelized
No Parallel processing with this module
dist = copy.deepcopy(clr_hac.dist)
n=len(vit_diar.unique('label'))
#print(dist)
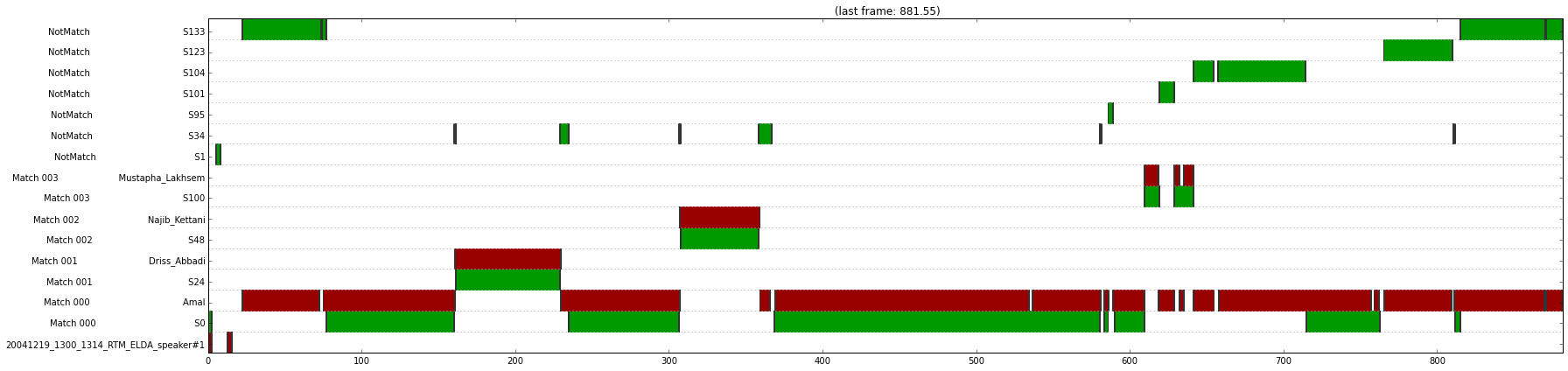
np.fill_diagonal(dist, np.min(dist))
# Plot the density map using nearest-neighbor interpolation
plot.figure(figsize=(10,10))
plot.pcolor(dist.T, cmap='RdBu')
plot.colorbar()
plot.show()
/Users/meignier/pyenv3/lib/python3.5/site-packages/matplotlib/collections.py:590: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
if self._edgecolors == str('face'):
clr_diar = clr_hac.perform(thr_clr, to_the_end=True)
if save_all:
clr_filename = os.path.join(wdir, show + '.clr.seg')
Diar.write_seg(clr_filename, clr_diar)
link, data = plot_dendrogram(clr_hac.merge, thr_clr, size=(25,6), log=True)
/Users/meignier/pyenv3/lib/python3.5/site-packages/matplotlib/collections.py:590: FutureWarning: elementwise comparison failed; returning scalar instead, but in the future will perform elementwise comparison
if self._edgecolors == str('face'):
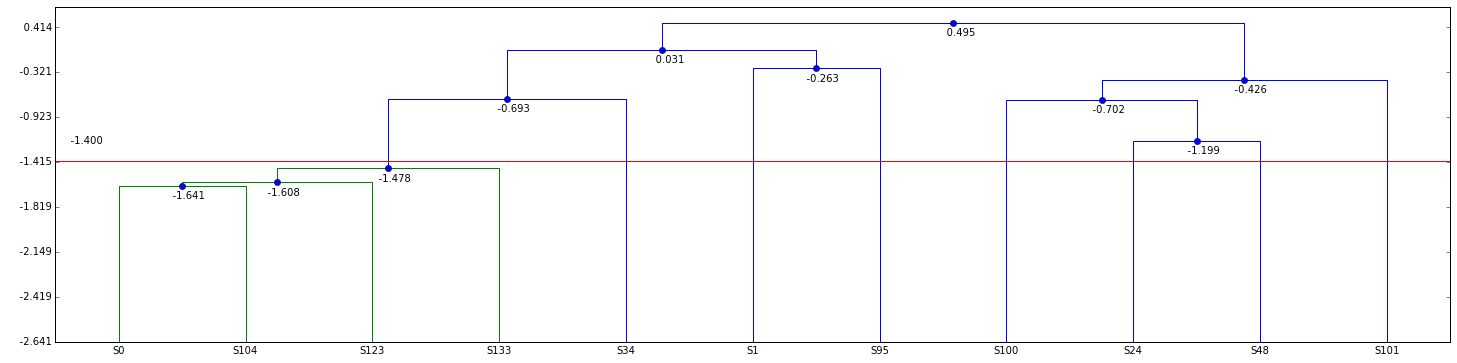
- Compute DER
der = DER(clr_diar, ref, uem, collar=25, no_overlap=False)
der.confusion()
der.assignment()
res = der.error()
print(show)
print(res.rate_header())
print(res.time())
print(res.rate())
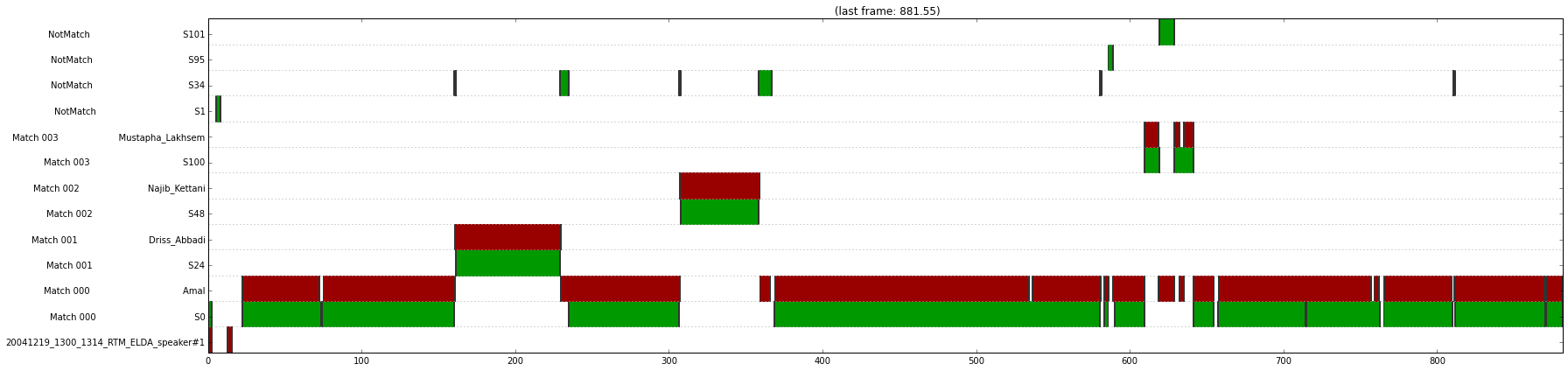
diff_diar = diar_diff(clr_diar, ref)
p = PlotDiar(diff_diar, size=(25, 6))
p.draw()
20041219_1300_1314_RTM_ELDA
|| sns fa miss | speaker fa miss conf ||
|| 1762.62s 12.49s 3.89s | 827.56s 12.49s 3.89s 25.94s ||
|| 0.93% 0.71% 0.22% | 5.11% 1.51% 0.47% 3.13% ||
uem from ref
append collar
/Users/meignier/pyenv3/lib/python3.5/site-packages/matplotlib/figure.py:1653: UserWarning: This figure includes Axes that are not compatible with tight_layout, so its results might be incorrect.
warnings.warn("This figure includes Axes that are not "
I-vector PLDA clustering¶
#matplotlib.use('TKAgg')
Graph and HAC¶
Licence¶
This file is part of S4D.
SD4 is a python package for speaker diarization based on SIDEKIT. S4D home page: http://www-lium.univ-lemans.fr/s4d/ SIDEKIT home page: http://www-lium.univ-lemans.fr/sidekit/
S4D is free software: you can redistribute it and/or modify it under the terms of the GNU Lesser General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
S4D is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License for more details.
You should have received a copy of the GNU Lesser General Public License along with SIDEKIT. If not, see http://www.gnu.org/licenses/.
Copyright 2014-2015 Sylvain Meignier